Method Overview
Site-directed mutagenesis (SDM) is a method to create specific, targeted changes in double stranded plasmid DNA. There are many reasons to make specific DNA alterations (insertions, deletions and substitutions), including:
- To study changes in protein activity that occur as a result of the DNA manipulation.
- To select or screen for mutations (at the DNA, RNA or protein level) that have a desired property
- To introduce or remove restriction endonuclease sites or tags
SDM is an in vitro procedure that uses custom designed oligonucleotide primers to confer a desired mutation in a double-stranded DNA plasmid. Formerly, a method pioneered by Kunkel (Kunkel, 1985) that takes advantage of a strain deficient in dUTPase and uracil deglycosylase so that the recipient E. coli degrades the uracil-containing wild-type DNA was widely used.
Currently, there are a number of commercially available kits that also require specific modification and/or unique E. coli strains (for example, the Phusion Site-Directed Mutagenesis from Thermo and the GeneArt system from Life). The most widely-used methods do not require any modifications or unique strains and incorporate mutations into the plasmid by inverse PCR with standard primers. For these methods, primers can be designed in either an overlapping (QuikChange, Agilent) or a back-to-back orientation (Q5 Site-Directed Mutagenesis Kit) (Figure 1).
Learn more about the Q5 Site-Directed Mutagensis Kit
![]() Use our free
Use our free
NEBaseChanger Tool!
Watch the NEBaseChanger Video Tutorial
Overlapping primer design results in a product that will re-circularize to form a doubly-nicked plasmid. Despite the presence of these nicks, this circular product can be directly transformed into E. coli, albeit at a lower efficiency than non-nicked plasmids. Back-to-back primer design methods not only have the advantage of transforming non-nicked plasmids, but also allow exponential amplification to generate significantly more of the desired product (Figure 2). In addition, because the primers do not overlap each other, deletions sizes are only limited by the plasmid and insertions are only limited by the constraints of modern primer synthesis. Currently, by splitting the insertion between the two primers, insertions up to 100 bp can routinely be created in one step using this method.
Before primers are designed, it is important to determine which mutagenesis workflow is to be used. Here we present a comparison of three commercially available kits (Figure 3) and a brief description of important features.
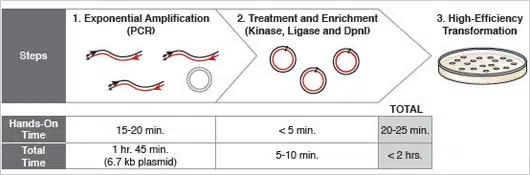
Figure 1: Site-specific mutagenesis proceeds in less than 2 hours.
The use of a master mix, a unique multi-enzyme KLD enzyme mix, and a fast polymerase ensures that, for most plasmids, the mutagenesis reaction is complete in less than two hours.
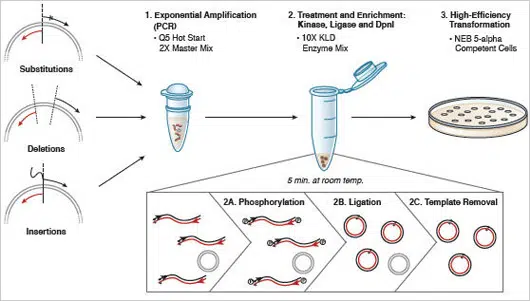
Figure 2: Q5 Site-Directed Mutagenesis Kit Overview.
This kit is designed for rapid and efficient incorporation of insertions, deletions and substitutions into doublestranded plasmid DNA. The first step is an exponential amplification using standard primers and a master mix fomulation of Q5 Hot Start High-Fidelity DNA Polymerase. The second step involves incubation with a unique enzyme mix containing a kinase, a ligase and DpnI. Together, these enzymes allow for rapid circularization of the PCR product and removal of the template DNA. The last step is a high-efficiency transformation into chemicallycompetent cells (provided).
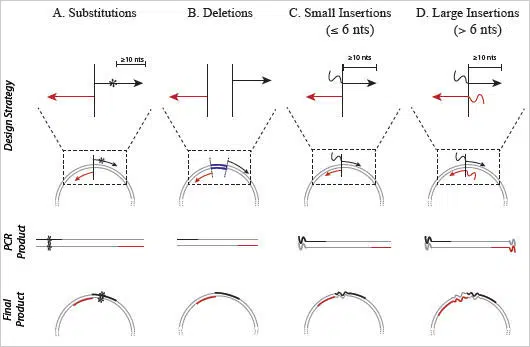
Figure 3: Primer Design for the Q5 Site-Directed Mutagenesis Kit
Substitutions, deletions and insertions are incorporated into plasmid DNA through the use of specifically designed forward (black) and reverse (red) primers. Unlike kits that rely on linear amplification, primers designed for the Q5 Site-Directed Mutagenesis Kit should not overlap to ensure that the benefits of exponential amplification are realized. A) Substitutions are created by incorporating the desired nucleotide change(s) (denoted by *) in the center of the forward primer, including at least 10 complementary nucleotides on the 3´side of the mutation(s). The reverse primer is designed so that the 5´ ends of the two primers anneal back-to- back. B) Deletions are engineered by designing standard, non-mutagenic forward and reverse primers that flank the region to be deleted. C) Insertions less than or equal to 6 nucleotides are incorporated into the 5´ end of the forward primer while the reverse primer anneals back-to-back with the 5´ end of the complementary region of the forward primer. D) Larger insertions can be created by incorporating half of the desired insertion into the 5´ ends of both primers. The maximum size of the insertion is largely dictated by oligonucleotide synthesis limitations.
Comparison to competition
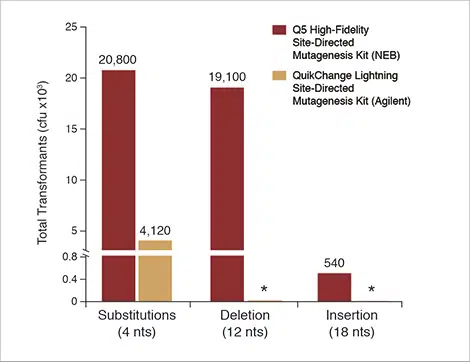
NEB’s Q5 SDM Kit delivers higher transformation efficiency than Agilent’s QuikChange® SDM Kit
Results from a substitution reaction (4 nt) using the back-to-back Control SDM Primer Mix and Control SDM Plasmid (6.7 kb) are shown, along with results from a 12 nt deletion experiment (5.8 kb plasmid) and an 18 nt insertion experiment (7.0 kb plasmid). In all three cases, over 90% of the resultant colonies had incorporated the desired mutation(s). Results are normalized to total transformants if cells were not diluted prior to plating. For comparison, the same substitution reaction (4 nt) was performed with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) following Agilent’s protocol and using Agilent’s primer design tool to design overlapping primers. *Note that the QuikChange kit does not accommodate deletions and insertions of this size, so no comparison could be made for these experiments.
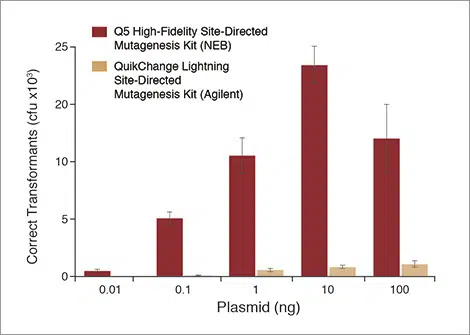
NEB’s Q5 SDM Kit produces a higher yield of correct transformants than the Agilent QuikChange Kit
The substitution reaction (4 nt) using the back-to-back Control SDM Primer Mix and Control SDM Plasmid (6.7 kb) was performed with various amounts of plasmid template as indicated. For comparison, the same experiment was performed with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) following Agilent’s protocol and using Agilent’s primer design tool to design overlapping primers. Results are normalized to total transformants if cells were not diluted prior to plating. Although successful experiments are possible outside this range, we recommend using between 0.1 and 100 ng of plasmid to achieve robust results.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.