Real-Time PCR
Real-Time PCR or quantitative PCR (qPCR) are methods used for the detection and quantification of nucleic acids (DNA or RNA).
There are two basic qPCR approaches:
Firstly, there are methods based on the incorporation of fluorescent dyes into double-stranded DNA. This is referred to as dye-based qPCR.
There are also various methods using fluorescence-labeled DNA probes. Consequently, this is referred to as probe-based qPCR.
Video: Overview of qPCR
Watch our video to get insights to the basics of real-time PCR, application areas and methods for the analysis of qPCR results.
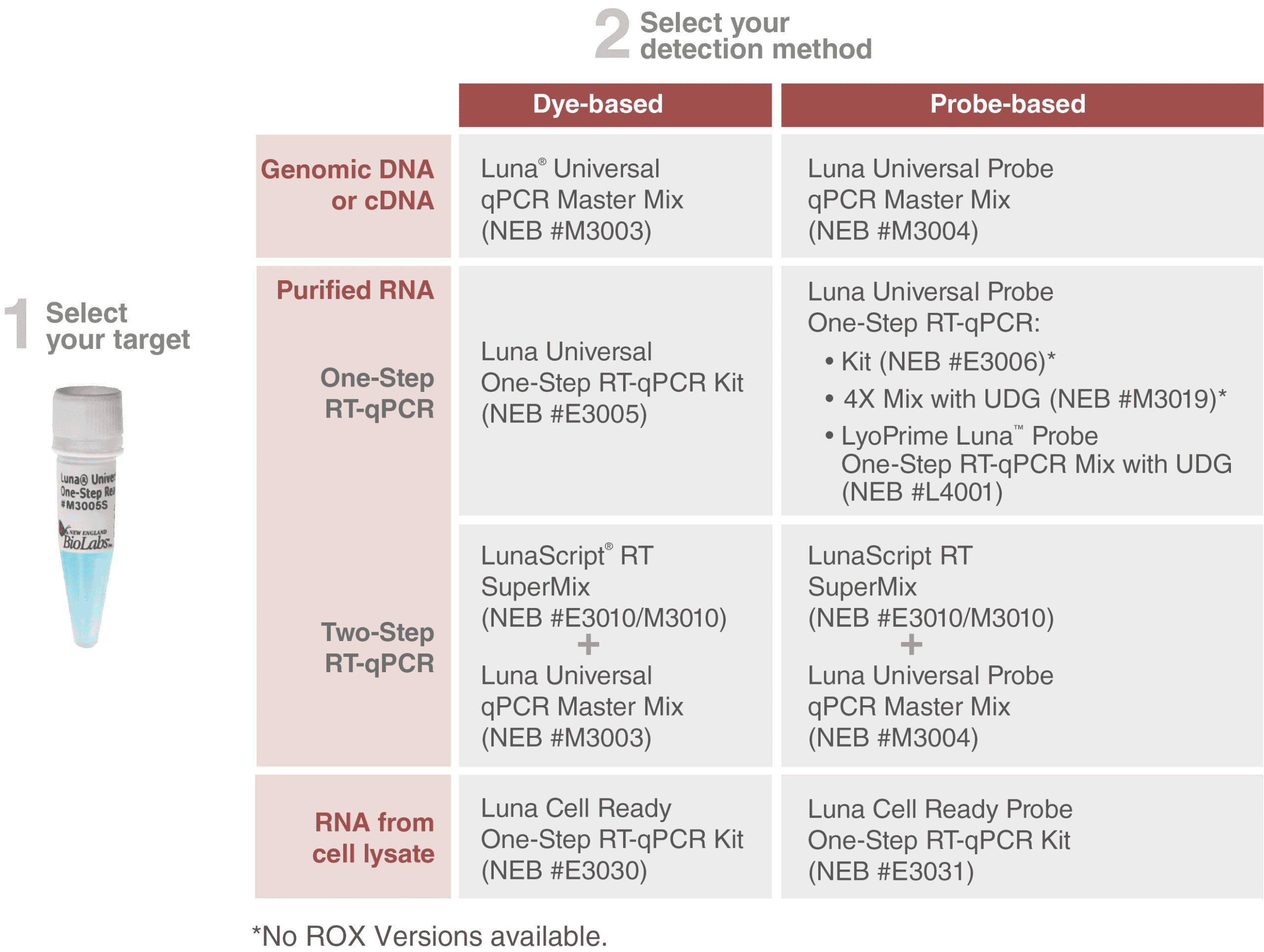
Information about our Luna qPCR and RT-qPCR products can be found here:
Dye-based qPCR
Dye-based real-time PCR methods utilize real time measurement of the fluorescence of a dye that specifically binds to double-stranded DNA (dsDNA) (e.g. SYBR® Green). The dye typically shows weak background fluorescence, which increases dramatically when binding to dsDNA. The amplification of the target sequence thus leads to an increase in fluorescence which is directly proportional to the amount of dsDNA present in each PCR cycle. This type of real-time PCR requires only two sequence-specific primers. The dye-based real-time PCR is therefore a fast and inexpensive way to examine a large number of samples.
A disadvantage of these dye-based methods is that they detect any dsDNA present in the reaction. This also includes non-specific products or primer dimers, which can lead to inaccurate quantification. Therefore, a denaturing / melting curve is usually created after each qPCR experiment to check the reaction specificity. This ensures that only the desired target has been amplified. Furthermore, in contrast to probe-based real-time PCR, which allows multiplexing, only one target sequence can be quantified in a dye-based qPCR experiment.
Convince yourself of the quality of our Luna Universal qPCR products:
Probe-based qPCR
Probe-based real-time PCR methods measure, also in real-time, the fluorescence of a fluorophore-labelled, sequence-specific probe. Probe-based qPCR is typically more specific than dye-based qPCR and is often the preferred technology for diagnostic applications.
Probe-based real-time PCR enables the user to quantify multiple targets in a single reaction (multiplexing) when a specific fluorophore is used for each target.
Probe designs can vary, the most common are so-called hydrolysis probes (e.g. TaqMan®). These include a 5′ reporter fluorophore and a 3′ quencher on a short oligonucleotide complementary to the target sequence. The fluorescence resonance energy transfer (FRET) between reporter and quencher prevents the emission of the fluorophore as long as the probe is intact (low fluorescence). During each PCR cycle, the 5′ flap endonuclease domain of the Taq DNA Polymerase hydrolyzes the probe while the primer is extended and the target sequence is amplified.
In addition to the hydrolysis probes, there are other probe designs such as molecular beacons, Scorpions® and dual hybridization probes.
Molecular beacons and Scorpions® are also based on the spatial separation of a reporter fluorophore and a quencher during PCR to release fluorescence.
Hybridization probes (LightCycler® probes) are based on FRET as well, but fluorescence is generated by energy transfer. Two sequence-dependent probes with compatible FRET dyes bind in close proximity to the target sequence. The excitation of the donor fluorophore, which is located at the 3′ end of the first probe, causes the emission of the acceptor fluorophore, which is located at the 5′ end of the second probe. When the target is amplified, the fluorescence signal of the acceptor fluorophore increases.
RT-qPCR
Real-Time PCR can also be used for the detection and quantification of RNA. A reverse transcriptase step is performed before the actual real-time PCR assay (RT-qPCR).
The reverse transcription can be performed separately from the PCR or directly in the qPCR mix. One-step RT-qPCR workflows are usually preferred in molecular diagnostic assays in which only one specific target has to be detected.
A separate cDNA synthesis with subsequent qPCR (Two-Step RT-qPCR) is used if several targets from the same starting material are to be detected or if archiving of the cDNA is required.
Our Luna qPCR Products:
As of: 01.01.2024
TaqMan® is a registered trademark of Roche Molecular Systems, Inc.
SYBR® is a registered trademark of Molecular Probes, Inc.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.