Genomic DNA Extraction and Purification
Quickly and easily purify high quality, high molecular weight gDNA from multiple sample types, all with one kit!
The Monarch Spin Genomic DNA Purification Kit is a comprehensive solution for cell lysis, RNA removal, and purification of intact genomic DNA (gDNA) from a wide variety of biological samples, including cultured cells, blood, and mammalian tissues. Additionally, bacteria and yeast can be processed with extra steps to enhance lysis in these tough-to-lyse samples. Protocols are also included to enable purification from clinically-relevant samples such as saliva and cheek swabs as well as rapid cleanup of previously extracted gDNA. The purified gDNA is suitable for downstream applications such as end-point PCR, qPCR and library prep for NGS. It typically has a peak size of > 50kb, making this kit an excellent choice upstream of long-read sequencing platforms.
The Monarch Spin Genomic DNA Purification Kit efficiently purifies high-quality, high molecular weight gDNA from a variety of sample types.
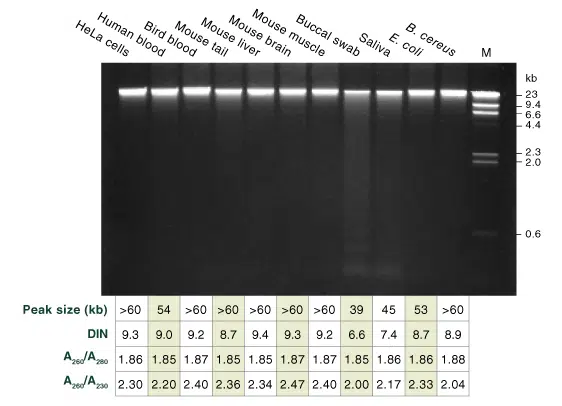
100 ng of genomic DNA from each sample was loaded on a 0.75% agarose gel. gDNA was isolated following the standard protocols for blood, cultured cells and tissue, and the supplemental protocols for buccal swabs, saliva, Gram– and Gram+ bacteria. Starting material used: 1 x 106 HeLa cells, 100 μl human blood, 10 μl bird blood, 10 mg frozen tissue powder, 1 buccal swab, 500 μl saliva and ~1 x 109 bacterial cells. Lambda DNA-Hind III digest (NEB #N3012) was used as a marker in the last lane (M). Purified gDNA samples were analyzed using a Genomic DNA ScreenTape® on an Agilent Technologies®4200 TapeStation®. Samples typically yield peak sizes 50–70 kb and DINs of ~9. The cell fractions processed in the buccal swab and saliva preps contain dead cells, as expected, causing a smear like pattern with typical low molecular weight apoptotic bands.
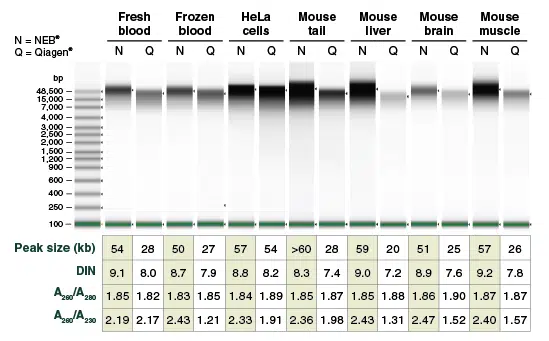
Agilent Technologies® 4200 TapeStation® Genomic DNA ScreenTape was used for analysis of blood, cultured cell, and tissue samples purified using the relevant protocols of the Monarch Genomic DNA Purification Kit and the Qiagen DNeasy Blood & Tissue Kit. gDNA was eluted in 100 μl and 1/100 of the eluates (~1 μl) was loaded on a Genomic DNA ScreenTape. Starting materials used: 100 μl fresh human whole blood, 100 μl frozen pig blood, 1×106 HeLa cells and 10 mg frozen tissue powder. Monarch-purified gDNA samples typically show peak sizes 50 – 70 kb and DINs of ~9. DNeasy-purified gDNA peak sizes are typically <30 kb with DINs ~7-8. DNeasy kits produce lower yields and low A260/A230 ratios for liver, brain, muscle and frozen blood.
Generate highly-intact genomic DNA suitable for long-range PCR, qPCR, NGS and other downstream applications.
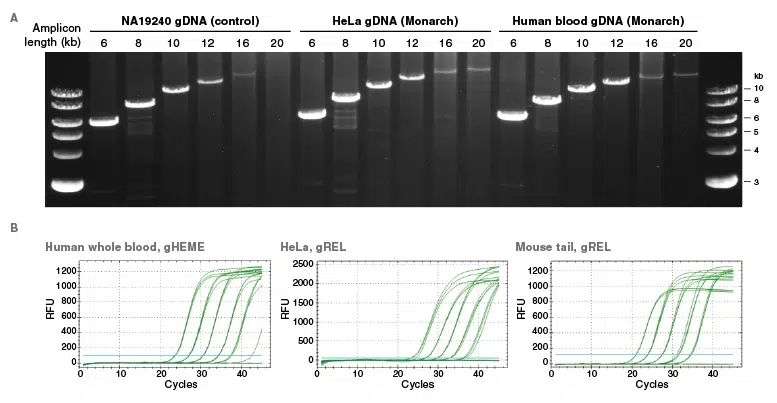
A. Amplification reactions were set up with primer pairs specific for 6, 8, 10, 12, 16 and 20 kb amplicons from human DNA. LongAmp®Hot Start Taq 2X Master Mix (NEB #M0533) was used and 25 ng template DNA was added to each sample. PCR reactions were carried out on an Applied Biosystems 2720 Thermal Cycler. Monarch-purified genomic DNA isolated from HeLa cells and human blood were compared to commercially available reference DNA from the human cell line NA19240 F11. 10 μl was loaded on a 1.5% agarose gel, using the 1 kb DNA Ladder (NEB #N3232) as a marker. Results indicated DNA was of high-integrity and suitable for long range PCR.
B. Monarch-purified genomic DNA from human whole blood, HeLa cells and mouse tail was diluted to produce a five log range of input template concentrations. The results were generated using primers targeting gHEME (human whole blood) and gREL (HeLa, mouse tail) for qPCR assays with the Luna® Universal qPCR Master Mix (NEB #M3003) and cycled on a Bio-Rad® CFX Touch qPCR thermal cycler. Results indicated that DNA is highly pure and free from inhibitors, optimal for qPCR.
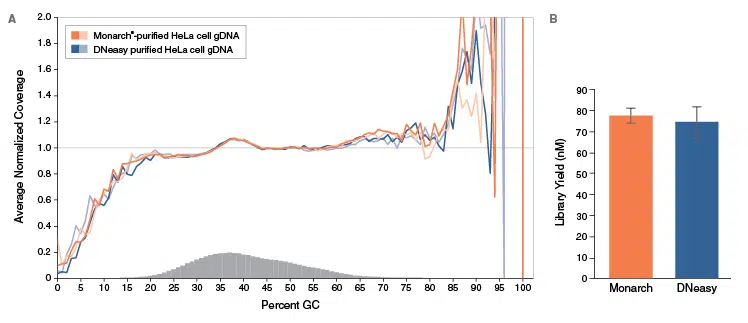
A. Duplicate libraries were made from 100 ng HeLa cell gDNA purified with Monarch (orange) or Qiagen DNeasy Mini Kit (blue) using the NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB #E7805). Libraries were sequenced on an Illumina MiSeq. Reads were mapped using Bowtie 2.2.4 and GC coverage was calculated using Picard’s CollectGCBiasMetrics (v1.117). Expected normalized coverage of 1.0 is indicated by the horizontal grey line, the number of 100 bp regions at each %GC is indicated by the vertical grey bars, and the colored lines represent the normalized coverage for each library. Monarch GC coverage matched Qiagen DNeasy results.
B. High yield libraries are achieved from Monarch-purified gDNA. Library yields of the samples described above were assessed on an Agilent Technologies® 2100 BioAnalyzer using a High Sensitivity DNA Kit.
Validated Sample Types:
- Mouse Tail
- Mouse Ear
- Mouse Liver
- Rat Liver
- Mouse Kidney
- Mouse Spleen
- Mouse Heart
- Mouse Lung
- Mouse Brain
- Rat Brain
- Mouse Muscle
- Rat Muscle
- Deer Muscle
- Human Blood
- Mouse Blood
- Rabbit Blood
- Pig Blood
- Guinea Pig Blood
- Cow Blood
- Horse Blood
- Dog Blood
- Chicken Blood
- HeLa Cells
- HEK293 Cells
- NIH3T3 Cells
- E. coli
- Rhodobacter sp.
- B. cereus
- T. kodakarensis
- S. cerevisiae
- Saliva
- Buccal swab
All Monarch nucleic acid kits are designed for minimal environmental impact
We achieve this by using particularly thin-walled columns and bottles. In this way we reduce the use of plastic and thus the amount of waste in the laboratory. In addition, all components of the packaging are made of recycled material.
Available Monarch gDNA products:
As of: 01.01.2024
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.