Engineered for performance under a wide range of conditions
For over 35 years, New England Biolabs has been developing innovative solutions for molecular biology applications. The respected leader in the field of restriction enzyme biology, NEB has developed a line of High-Fidelity (HF®) Restriction Enzymes. These engineered enzymes have the same specificity as the native enzyme, with the added benefit of reduced star activity, rapid digestion (5-15 minutes) and 100% activity in CutSmart® Buffer.
Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.
Optimized performance for a wide range of conditions
High Fidelity Restriction Enzymes have been engineered by exchanging functional amino acid residues and then screening for optimal mutants that perform under a wide range of conditions. Whether you are setting up digests for 5-15 minutes or overnight, or using varying amounts of enzymes, HF enzymes ensure the performance you need.
Advantages
![]()
Reduced star activity eliminates unwanted cleavage
![]()
One buffer convenience with no loss of performance
![]()
Time-Saver qualified for 5-15 minute digests, flexible enough to digest overnight.
![]() Added flexibility without added cost
Added flexibility without added cost
![]() Including 1 ml Gel Loading Dye, Purple (6X) gratis
Including 1 ml Gel Loading Dye, Purple (6X) gratis

Available HF Enzymes
|
|
|
Star Activity
Under non-standard reaction conditions, some restriction enzymes are capable of cleaving sequences which are similar, but not identical, to their defined recognition sequence. This altered specificity has been termed “star activity”. It has been suggested that star activity is a general property of restriction endonucleases (1) and that any restriction endonuclease will cleave noncanonical sites under certain extreme conditions, some of which are listed below. Although the propensity for star activity varies, the vast majority of enzymes from New England Biolabs will not exhibit star activity when used under recommended conditions in their supplied NEBuffers. If an enzyme has been reported to exhibit star activity, it will be indicated in the product entry found in the catalog, on the datacard or on our web site.
The manner in which an enzyme’s specificity is altered depends on the enzyme and the reaction conditions which induce star activity. The most common types of altered activity are single base substitutions, truncation of the outer bases in the recognition sequence, and single-strand nicking (2). Some enzymes exhibit relaxation of sequence specificity under standard conditions and in the presence of the cognate site are capable of cleaving non-cognate (secondary) sites (3).
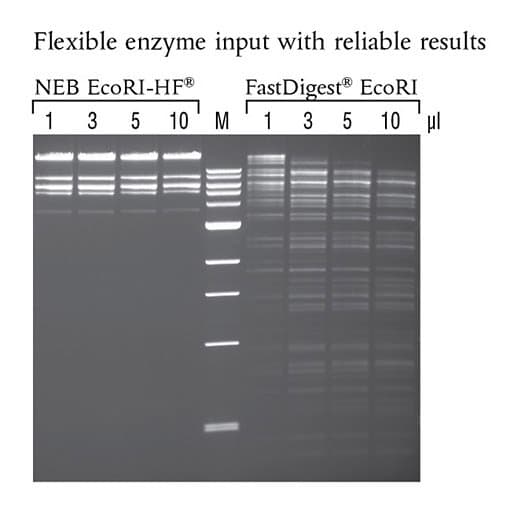 EcoRI-HF® (NEB #R3101) shows no star activity in overnight digests, even when used at higher concentrations. 50 μl reactions were set up using 1 μg of Lambda DNA, the indicated amount of enzyme and the recommended reaction buffer. Reactions were incubated overnight at 37°C. Marker M is the 1 kb DNA Ladder (NEB# N3232)
EcoRI-HF® (NEB #R3101) shows no star activity in overnight digests, even when used at higher concentrations. 50 μl reactions were set up using 1 μg of Lambda DNA, the indicated amount of enzyme and the recommended reaction buffer. Reactions were incubated overnight at 37°C. Marker M is the 1 kb DNA Ladder (NEB# N3232)
How to avoid Star Activity
| Conditions that Contribute to Star Activity | Steps that can be Taken to Inhibit Star Activity |
|---|---|
| High glycerol concentration (> 5% v/v) | Restriction enzymes are stored in 50% glycerol, therefore the amount of enzyme added should not exceed 10% of the total reaction volume. Use the standard 50 µl reaction volume to reduce evaporation during incubation. |
| High concentration of enzyme/µg of DNA ratio (varies with each enzyme, usually 100 units/µg) | Use the fewest units possible to achieve digestion. This avoids overdigestion and reduces the final glycerol concentration in the reaction. |
| Non-optimal buffer | Whenever possible, set up reactions in the recommended buffer. Buffers with differing ionic strength and pH may contribute to star activity. |
| Prolonged reaction time | Use the minimum reaction time required for complete digestion. Prolonged incubation may result in increased star activity, as well as evaporation. |
| Presence of organic solvents [DMSO, ethanol (4), ethylene glycol, dimethylacetamide, dimethylformamide, sulphalane (5)] | Make sure the reaction is free of any organic solvents, such as alcohols, which might be present in the DNA preparation. |
| Substitution of Mg2+ with other divalent cations (Mn2+, Cu2+, Co2+, Zn2+) | Use Mg2+ as the divalent cation. Other divalent cations may not fit correctly into the active site of the restriction enzyme, possibly interfering with proper recognition. |
References:
1. Nasri, M. and Thomas, D. (1986) Nucleic Acids Res. 14, 811. PMID: 3003698
2. Barany, F. (1988) Gene 65, 149. PMID: 2842230
3. Bitinaite, J. and Schildkraut, I. (2002) Proc. Natl. Acad. Sci USA 99, 1164-1169. PMID: 11818524
4. Nasri, M. and Thomas, D. (1987) Nucleic Acids Res. 15, 7677. PMID: 2823216
5. Tikchinenko, T. I. et al. (1978) Gene. 4, 195-212. PMID: 33871
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.

