EpiMark Methylated DNA Enrichment Kit
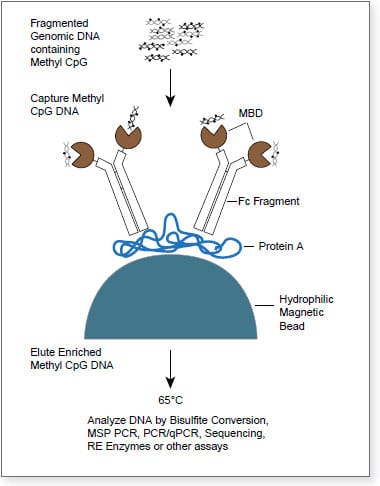
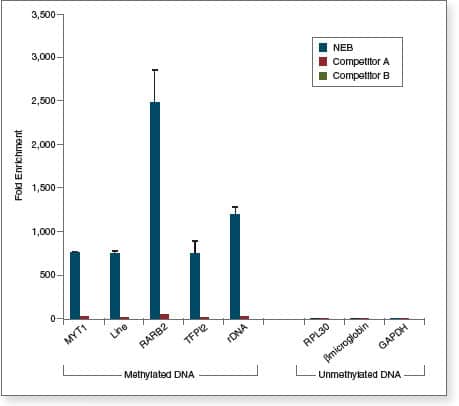
The EpiMark® Methylated DNA Enrichment Kit enables the enrichment of double-stranded CpG methylated DNA based on CpG methylation density. It utilizes the methyl-CpG binding domain of human MBD2a protein as a capture agent. The protein is fused to the Fc tail of human IgG1 (MBD2a-Fc), which is coupled to Protein A Magnetic Beads (MBD2a-Fc/Protein A Bead). This stable complex will selectively bind double-stranded methylated CpG containing DNA. The high binding affinity of the beads coupled with optimized reagents increases sensitivity and accuracy. This kit contains all the individual components necessary to achieve enrichment in less than two hours using a four step process.
Advantages:
- Increased sensitivity
- Easy-to-use protocol yields enriched methylated DNA in less than 2 hours
- Amenable to downstream applications, including next generation sequencing
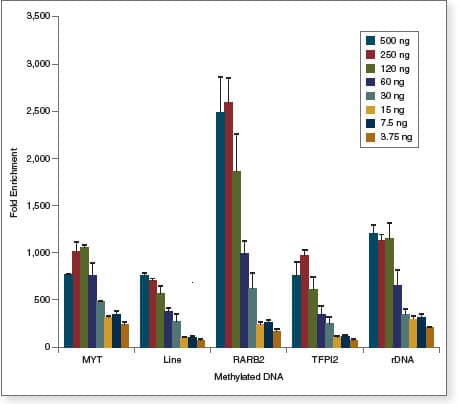
- Suitable for low levels of input DNA
Use directly in downstream analysis:
- Endpoint PCR
- Real-time PCR
- Bisulfite conversion
- Cloning and sequencing
- Direct sequencing
- Library preparation for high throughput sequencing
- Labelling for DNA microarray analysis
- Methylation-sensitive restriction enzyme-based assays
EpiMark 5-mC and 5-hmC Analysis Kit
Breakthrough in 5-hmC Quantitation for Epigenetics
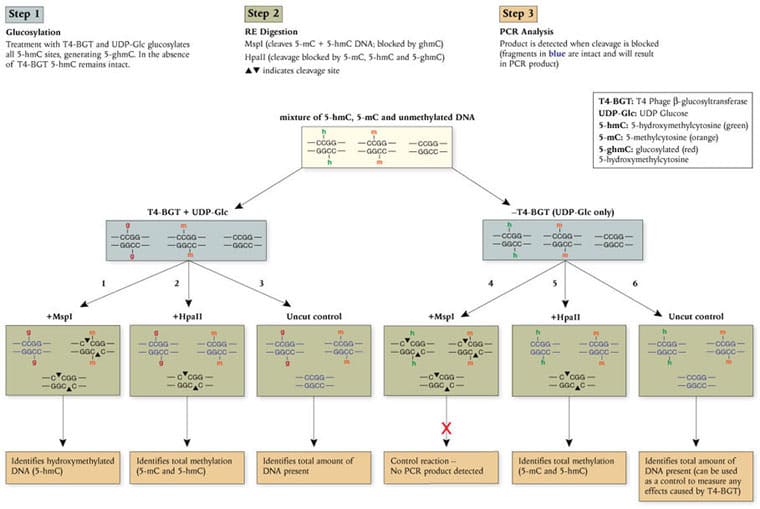
New England Biolabs is pleased to introduce the EpiMark® 5-hmC and 5-mC Analysis Kit, a simple and robust method for the identification and quantitation of 5-hydroxymethylcytosine (5-hmC) and 5-methylcytosine (5-mC) within a specific DNA locus. This enzymatic approach utilizes the differential methylation sensitivity of the isoschizomers MspI and HpaII in a simple 3-step protocol (described below). The EpiMark Kit is the first commercially available PCR-based assay to reproducibly identify and quantitate the presence of 5-hmC and 5-mC, and expands the potential for new biomarker discovery.
Advantages:
- Reproducible quantitation of 5-hmC and 5-mC
- Easy-to-use protocols
- Compatible with existing techniques (PCR)
- Amenable to high throughpu
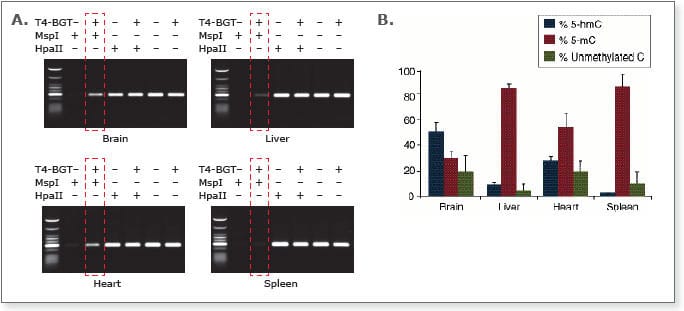
Analysis of the different methylation states in Balb/C mouse tissue samples using the EpiMark 5-hmC and 5-mC Analysis Kit.
A) Endpoint PCR of the 6 different reactions needed for methylation analysis. The boxed lanes indicate the presence of 5-hmC.
B) Real time PCR data was used to determine amounts of 5-hmC and 5-mC present. The results demonstrate a variation in 5-hmC levels in the tissue sources indicated.
EpiMark Bisulfite Conversion Kit
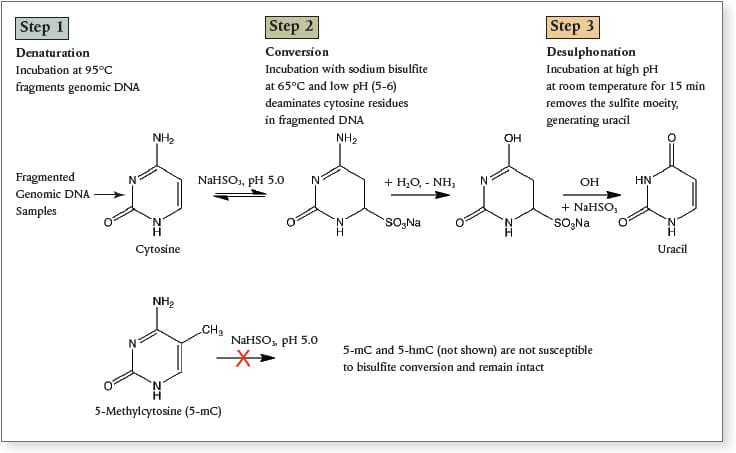
Sodium bisulfite conversion is the current “gold standard” in methylation analysis. This technique can reveal the methylation status of every cytosine residue, and it is amenable to massively parallel sequencing methods.
Bisulfite conversion, involves the conversion of unmodified cytosines to uracil, leaving the modified bases (5-mC and 5-hmC). The EpiMark® Bisulfite Conversion Kit is designed for the detection of methylated cytosine, using a series of alternating cycles of thermal denaturation, followed by incubation with sodium bisulfite. This kit includes all the reagents necessary for complete bisulfite conversion, including spin columns. Amplification of bisulfite-treated samples can then be performed using EpiMark® Hot Start Taq DNA Polymerase.
Advantages
- Complete conversion of unmodified cytosines to uracil
- Easy-to-use protocol ensures reliable and consistent results
- All reagents, including purification columns, are provided
Overview of bisulfite conversion
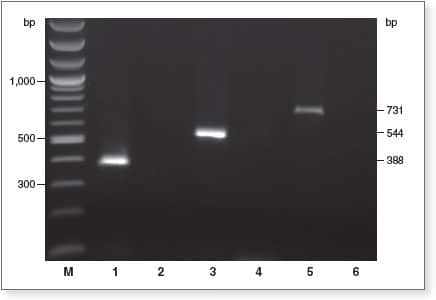
EpiMark Kit enables complete DNA conversion
1 µg of genomic DNA was bisulfite-treated using the Epimark Bisulfite Conversion Kit, and 2 µl of eluted DNA from 40 µl of elutant was analyzed by end-point PCR using EpiMark Hot Start Taq. Amplification with primer pairs for bisulfite converted DNA (lanes 1, 3, and 5), or with primer pairs for unconverted DNA (lanes 2, 4, and 6) were performed; lanes 2, 4, and 6 show no amplification product, indicating complete conversion.
EpiMark Hot Start DNA Polymerase
EpiMark® Hot Start Taq DNA
Polymerase – an excellent choice for use on bisulfite-treated DNA
EpiMark Hot Start Taq DNA Polymerase is a mixture of Taq DNA Polymerase and a temperature sensitive, aptamer-based inhibitor. This inhibitor binds reversibly to the enzyme, inhibiting polymerase activity below 45°C, but releases the enzyme during normal PCR cycling conditions. This permits PCR reactions to be assembled at room temperature and eliminates an activation step. This aptamer-based hot start activity combined with the supplied reaction buffer, that has been optimized for amplification of converted DNA, makes EpiMark Hot Start Taq an excellent choice for use on bisulfite-treated DNA.
EpiMark Nucleosome Assembly Kit
In eukaryotes, chromatin is organized into nucleosome core particles (NCPs) that consist of approximately 147 bp of DNA and an octamer complex made up of two molecules of each histone (H2A, H2B, H3 and H4). The linker histone, H1, can further condense chromatin by binding to linker DNA between the nucleosome core particles. For histone modification studies, New England Biolabs offers a selection of unmodified, recombinant human histones that function as substrates and can assemble into octamers. Octamer formation can be further simplified using the EpiMark Nucleosome Assembly Kit which contains preformed Histone H2A/H2B Dimer and Histone H3.1/H4 Tetramer.
Applications:
- Purification and characterization of enzymes that modify histone proteins
- Octamer modification studies
- Carrier chromatin immunoprecipitation (CChIP)
The EpiMark Nucleosome Assembly Kit contains all the components necessary for recombinant human nucleosome formation with user-supplied or the provided control DNA. These recombinant nucleosomes may serve as better substrates for enzymes (e.g. acetylases, methylases, sumoylases) or other chromatin interacting proteins that are unable to bind and/or modify individual histones or DNA alone. Additionally, the Histone H2A/H2B Dimer and the Histone H3.1/H4 Tetramer may themselves contain the required fold or structure to serve as more favorable substrates than the individual histones.
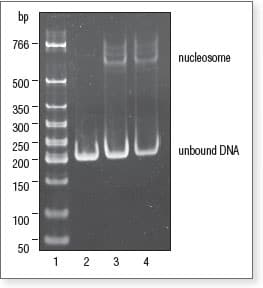
Gel shift assay to visualize nucleosome assembly
Samples from nucleosome assembly reactions were run on 6% polyacrylamide gel in 0.5X TBE.
Lane 1: Low Molecular Weight DNA Ladder (NEB #N3233)
Lane 2: Nucleosome Control DNA
Lane 3: 0.5:1 ratio of Octamer* to DNA
Lane 4: 1:1 ratio of Octamer* to DNA
*Octamer = 2:1 mix of Histone H2A/H2B Dimer and Histone H3.1/H4 Tetramer.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.