The K. lactis Expression Kit provides an easy method for expressing a gene of interest in the yeast Kluyveromyces lactis.
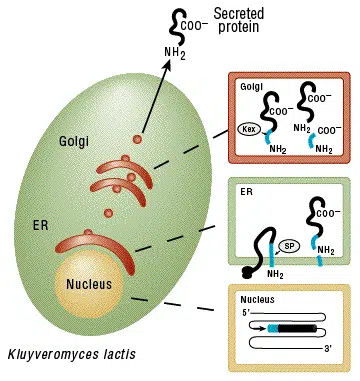
Proteins may be produced intracellularly or be secreted using the supplied integrative expression vector pKLAC2. To achieve protein secretion, a gene of interest is cloned downstream of the K. lactis α-mating factor secretion domain (α-MF) which is eventually processed in the Golgi resulting in secretion of the desired protein.
- Supplied with competent K. lactis cells (NEB# C1001)
- Allows cloning and expression of genes toxic to E. coli
- Rapid high cell density growth results in high yield protein expression
- Easy-to-use protocols for those inexperienced with yeast systems
- No expensive antibiotics or methanol required
- Attractive commercial sub-licensing
The K. lactis expression system offers several advantages over other yeast and bacterial protein expression systems:
First, K. lactis has been used to produce proteins at industrial scale in the food industry for over a decade due to its ability to rapidly achieve high culture densities and abundantly produce recombinant proteins.
Second, yeast expression is driven by a variant of the strong LAC4 promoter that has been modified to lack background expression in E. coli. Therefore, genes toxic to E. coli can be cloned into pKLAC2 in bacteria prior to their expression in yeast.
Third, the kit includes highly competent K. lactis cells making the technology easy-to-use for those not accustomed to working with yeast. Their high transformation efficiency makes the system suitable for methods that require large numbers of transformants, for example, expression cloning using cDNA libraries. Selection of yeast transformants uses a unique antibiotic-free method in which acetamidase (amdS) expressed from pKLAC2 permits transformed cells to utilize acetamide as a sole nitrogen source on defined medium. Acetamide selection promotes formation of cells containing multiple integrations of pKLAC2 which results in higher yields of protein.
Finally, proteins expressed in K. lactis have access to eukaryotic protein folding and glycosylation machinery that E. coli cells do not possess, making it an important alternative to bacterial expression systems.
In the nucleus, an integrated expression vector encoding a fusion between the α-MF domain (blue) and a desired protein (black) is expressed. A signal peptide in the α-MF domain directs entry of the fusion protein into the endoplasmic reticulum (ER) and is removed by signal peptidase (SP). The fusion protein is transported to the Golgi where the Kex protease removes the α-MF domain. The protein of interest is then secreted from the cell.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.

